
These differences in the microenvironments of the AF and NP compartments may influence their gene signatures. The environment within the NP is largely hypoxic, and the cells of the NP rely on diffusion of oxygen and nutrients through the endplate 9, 11. The IVD is predominantly avascular, with vasculature limited to the cartilaginous endplate and outer annulus fibrosus 6, 9, 10. These metabolic changes can alter the composition of the extracellular matrix (ECM) in the NP and AF compartments 1, 8 that may, in turn, affect the load-bearing and mechanical properties of the IVD leading to degeneration. It is postulated that catabolic and anabolic dysregulation of the cells within the AF and NP compartments play a pivotal role in the pathobiology of IDD 1, 7. There are three distinct anatomical regions in IVD- an outer annulus fibrosus (AF) composed of radially arranged collagen filaments that enclose an inner gelatinous nucleus pulposus (NP), as well as a cartilaginous endplate that demarcates the boundary between the IVD and the bony vertebral body 3, 5, 6. IDD has a significant economic burden, amounting to over one hundred billion dollars annually in direct and associated costs 2. Intervertebral disc (IVD) degeneration (IDD) is a pathophysiological process and is a common contributor to the development of chronic lower back pain 1, 2, 3, 4. Our regulatory network analysis identified FOXM1 and KDM4E as signature transcription factor of AF and NP respectively, which might be involved in the regulation of core genes of AF and NP transcriptome. Subsequent interaction network analysis revealed a structured network of extracellular matrix genes in NP compartments. Further, functional annotation clustering analysis revealed the enrichment of receptor signaling pathways genes in AF cells, while NP cells showed high expression of genes related to the protein synthesis machinery. Gene enrichment analysis showed that SFRP1, BIRC5, CYTL1, ESM1 and CCNB2 genes were highly expressed in the AF cells whereas, COL2A1, DSC3, COL9A3, COL11A1, and ANGPTL7 were mostly expressed in the NP cells. Our systematic and comprehensive analyses revealed distinct genetic architecture of human NP and AF compartments and identified 2,196 differentially expressed genes. We defined the transcriptome map of healthy human IVD by performing single-cell RNA-sequencing (scRNA-seq) in primary AF and NP cells isolated from non-degenerated lumbar disc.

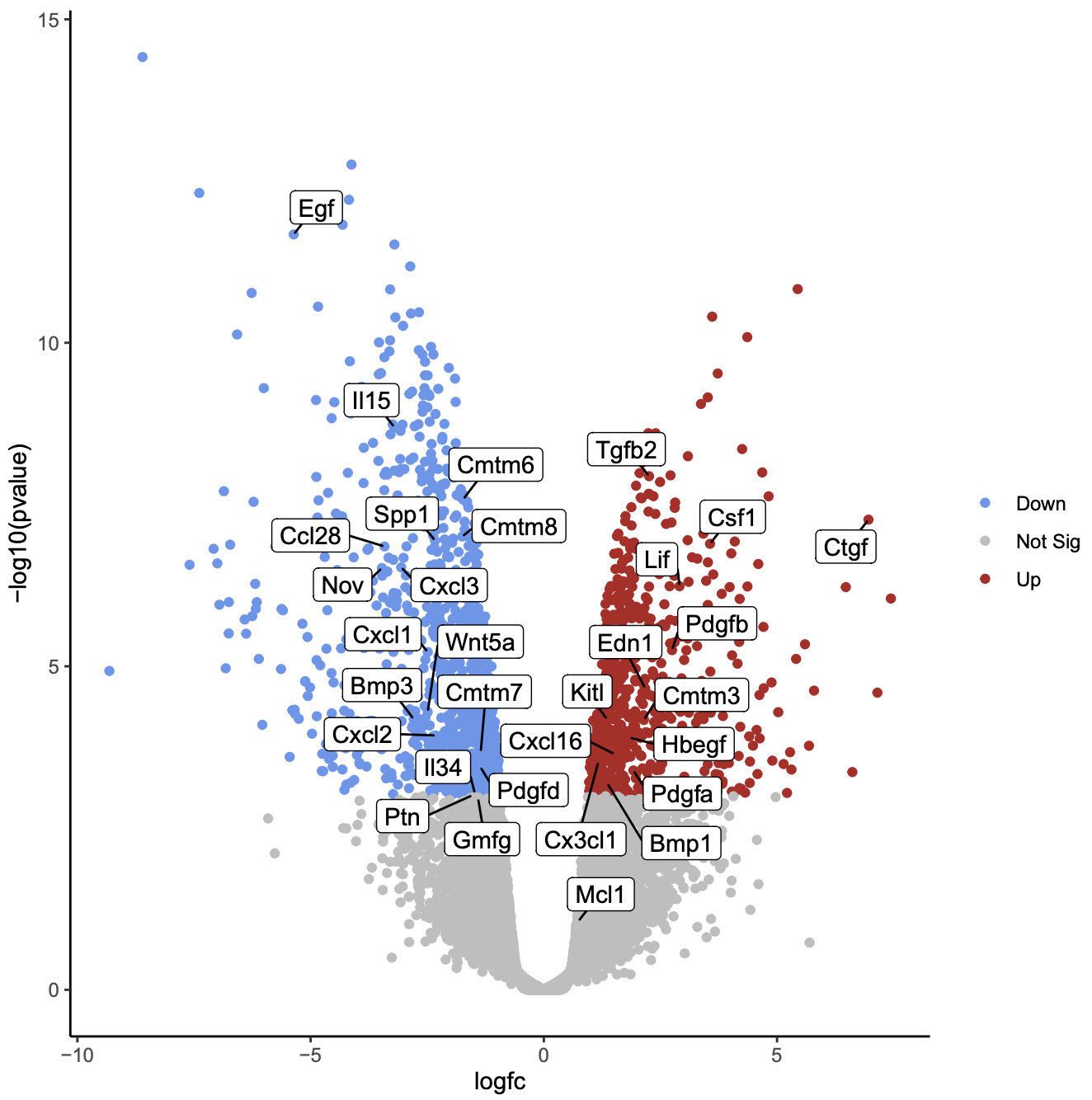
While various aspects of IDD progression have been reported, the underlying molecular pathways and transcriptional networks that govern the maintenance of healthy nucleus pulposus (NP) and annulus fibrosus (AF) have not been fully elucidated. Intervertebral disc (IVD) disease (IDD) is a complex, multifactorial disease.


 0 kommentar(er)
0 kommentar(er)
